library(slendr)
init_env()
simulate_afs <- function(Ne) {
# In here you should write code which will:
# 1. create one population with a given Ne (provided as a function argument)
# 2. compile a model using `simulation_length =` and `generation_time =`
# 3. simulate a tree sequence
# 4. select names of 10 samples (doesn't matter which, "pop_1", "po2_", ...)
# 5. compute AFS vector from those 10 individuals using `ts_afs()`
# `result` is a variable with your 10-sample AFS vector (we remove the
# first element because it's not meaningful for our example)
return(result[-1])
}
afs_1 <- simulate_afs(Ne = 1000) # simulate AFS from a Ne = 1000 model...
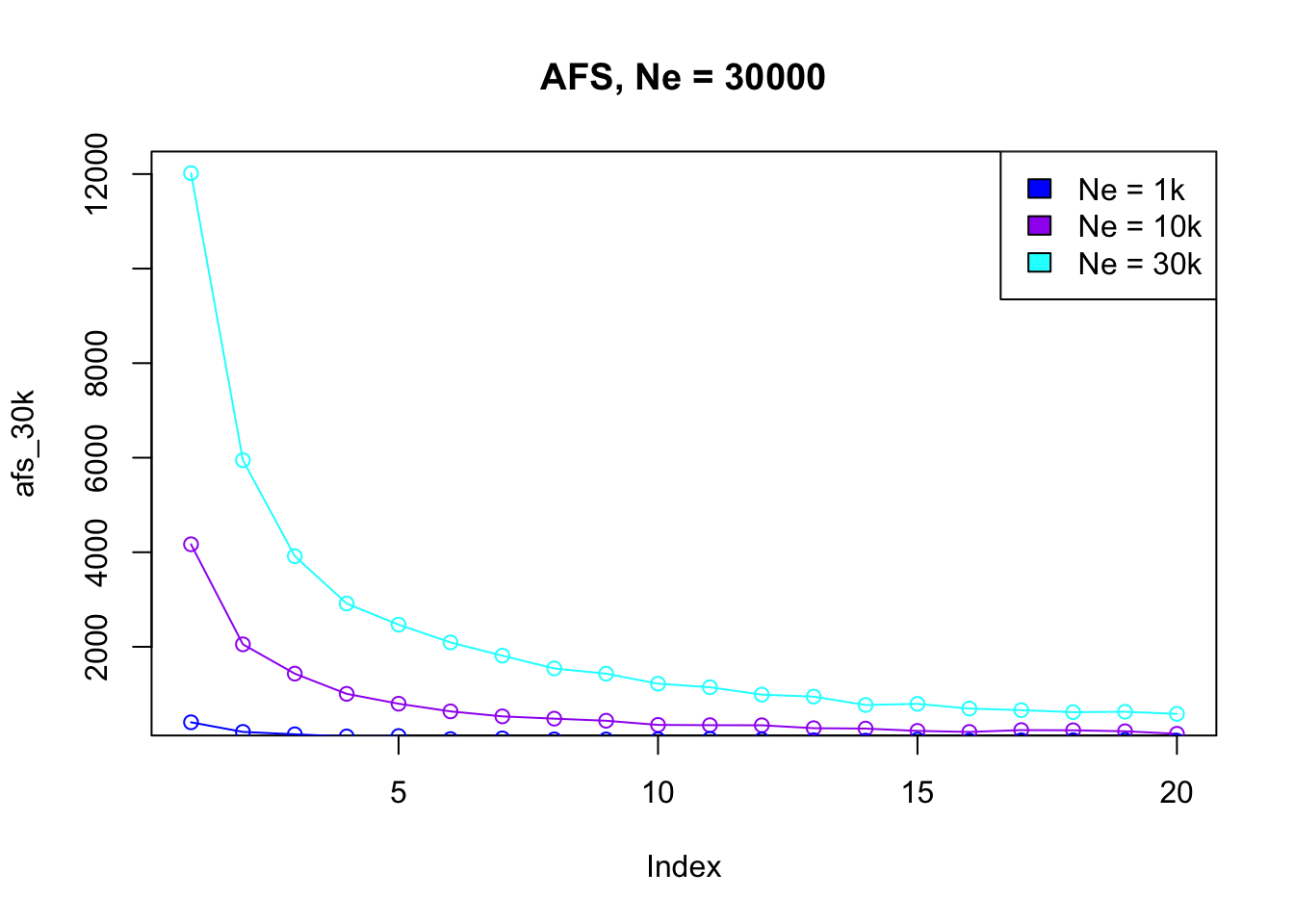
plot(afs_1, type ="o") # ... and plot it\(N_e\) inference with AFS
So far we’ve learned how slendr provides an easy way to define demographic models in R and simulate (even very large!) tree sequences from them. This allows us to quickly verify our intuition about some popgen problem (things like “Hmmm, I wonder what would an \(f_4\) statistic look like if my model includes this particular gene-flow event?), in just a few lines of R. There have been instances in which we’ve been able to even answer questions like this directly in a meeting, pretty much on the spot! This makes slendr a very powerful “popgen calculator”.
Now let’s take things one step further. Imagine you gathered some empirical data, like an allele frequency spectrum (AFS) from a population that you study. That data was, in the real world, produced by some (hidden) biological process (demographic history) that we want to learn about. For instance, the population we study had some \(N_e\), which we don’t know the value of (the only thing we have is the observed AFS) but we want to infer that value.
Simulations can be a great tool to estimate the most likely value of such an unknown parameter. Briefly speaking, in this particular toy example, we can simulate a large number of AFS vectors (each resulting from a different assumed \(N_e\) value) and then pick just those \(N_e\) values (or just one \(N_e\) value) which produced a simulated AFS closest to the observed AFS.
This is exactly what you’ll be doing just now.
Exercise 1: A self-contained slendr function of \(N_e \rightarrow \textrm{AFS}\)
In a new script inference-afs.R write a custom R function called simulate_afs(), which will take Ne as its only parameter. Use this function to compute (and return) AFS vectors for a couple of Ne values of your choosing, but staying between Ne = 1000 and Ne = 30000 Plot those AFS vectors and observe how (and why?) do they differ based on Ne parameter you used in each respective simulation.
Hint: The function should create a one-population forward-time model (our population starting at time = 1, with the model simulation_length = 100000 and generation_time = 1 in compile_model()), simulate 10Mb tree sequence using msprime() (recombination rate 1e-8) and then overlay neutral mutations on it at mutation_rate = 1e-8), compute AFS for 10 samples and return the AFS vector as result of this custom function.
Hint: If you’ve never programmed before, the concept of a “custom function” might be very alien to you. Again, if you need help, feel free to start building your inference-afs.R solution based on this “template” (just fill in missing relevant bits of slendr code that you should be already familiar with):
Note: Remember that you should drop the first element of the AFS vector produced by ts_afs() (for instance with something like result[-1] if result contains the output of ts_afs()) technical reasons related to tskit. You don’t have to worry about that here, but you can read this for more detail.
Hint: If the above still doesn’t make any sense to you, feel free to copy-paste the function from the solution below into your script and work with that function instead!
When used in R, your custom function should work like this (the simulation is stochastic, so your numbers will be different, of course):
# This gives us a vector of singletons, doubletons, etc., etc., all the way
# to the number of fixed mutations in our sample of 10 individuals
simulate_afs(Ne = 1000) [1] 382 201 126 102 74 44 48 60 32 35 29 32 30 21 22 26 40 36 21
[20] 34Exercise 2: Estimating unknown \(N_e\) from empirical AFS
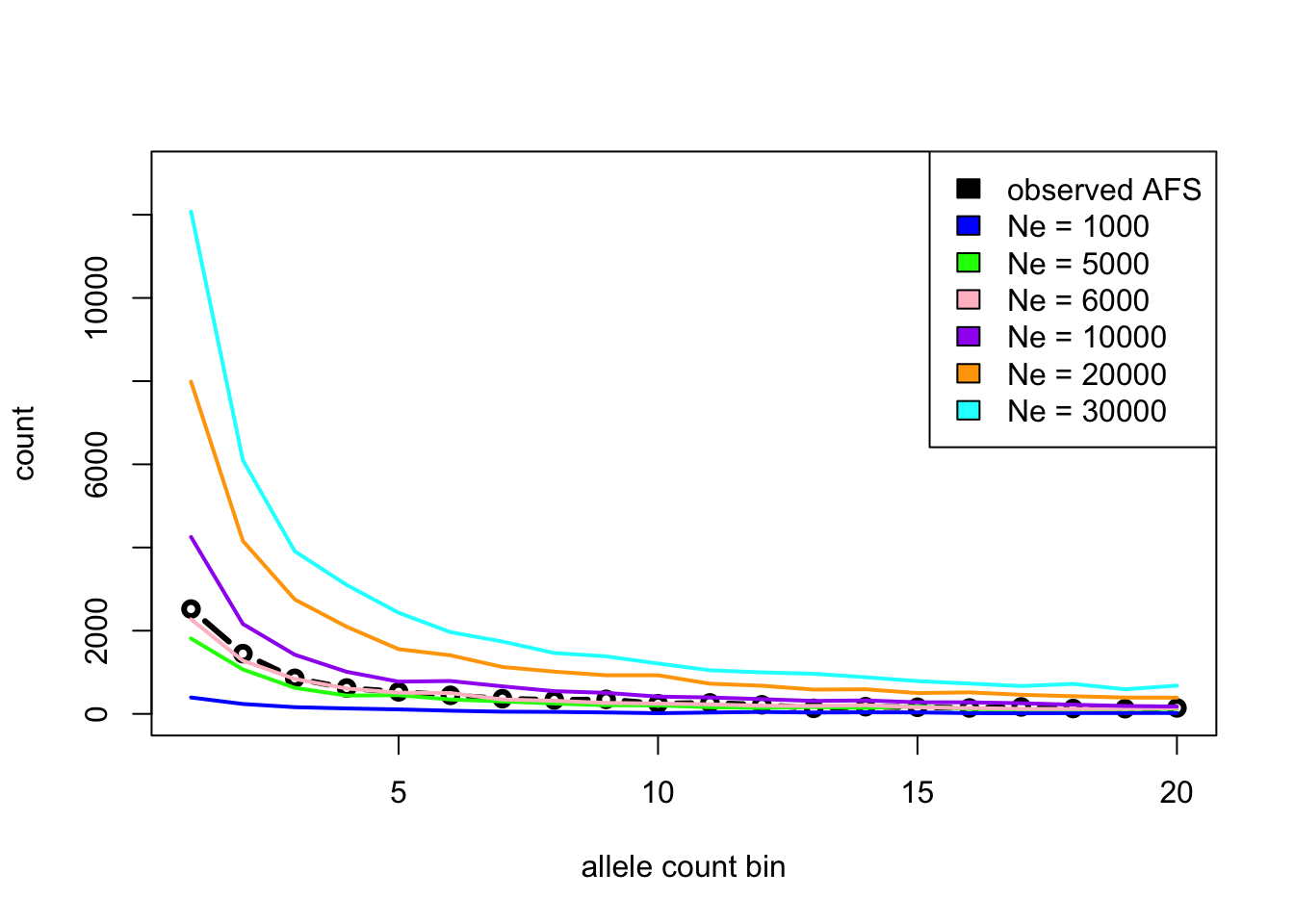
Imagine you sequenced 10 samples from a population and computed the following AFS vector (which contains, sequentially, the number of singletons, doubletons, etc., in your sample from a population):
afs_observed <- c(2520, 1449, 855, 622, 530, 446, 365, 334, 349, 244,
264, 218, 133, 173, 159, 142, 167, 129, 125, 143)You know (maybe from some fossil evidence) that the population probably had a constant \(N_e\) somewhere between 1000 and 30000 for the past 100,000 generations, and had mutation and recombination rates of 1e-8 (i.e., parameters already implemented by your simulate_afs() function – how convenient!).
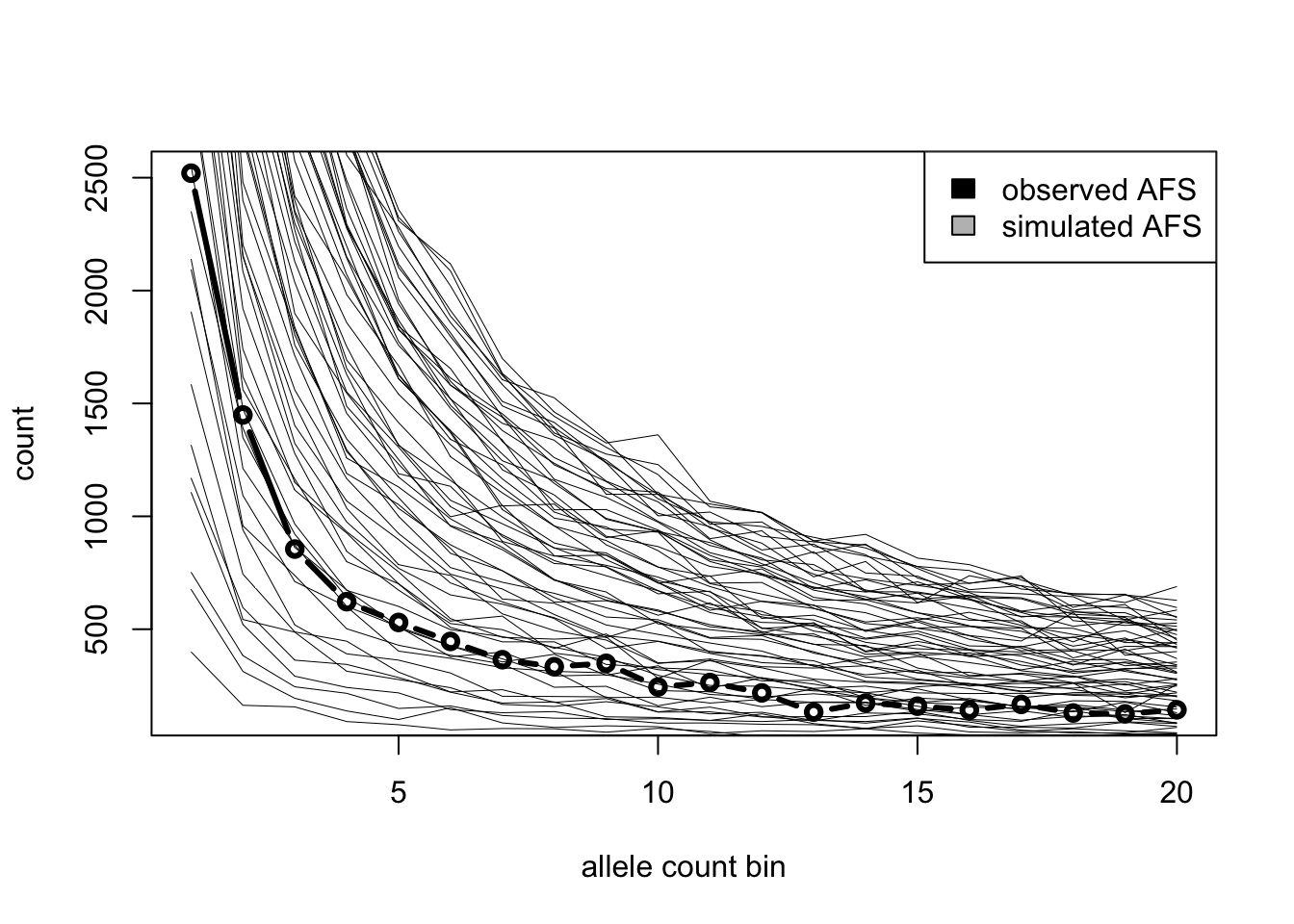
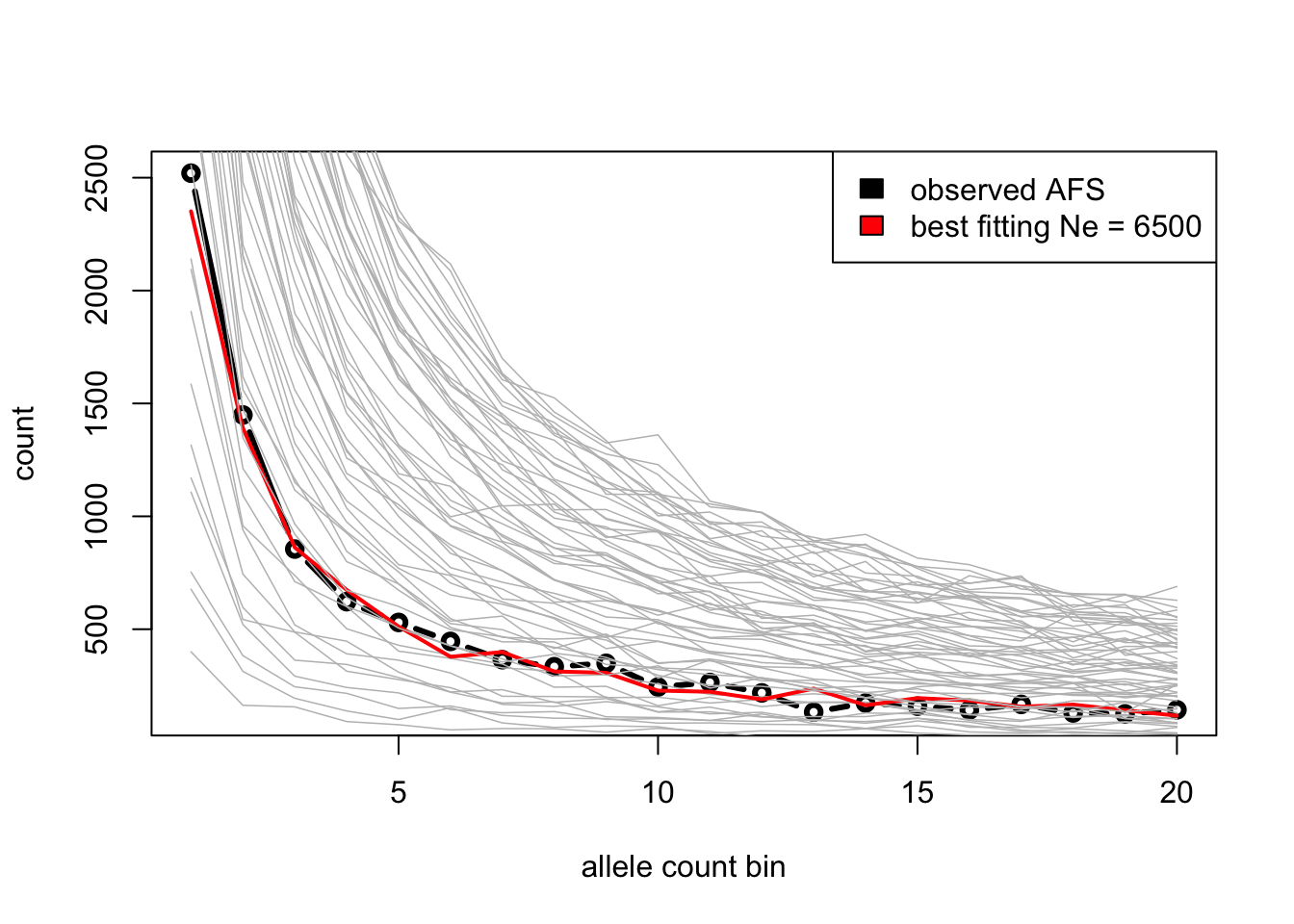
Use slendr simulations to guess the true (and hidden!) \(N_e\) given the observed AFS by running simulations for a range of \(N_e\) values and finding out which \(N_e\) produces the closest AFS vector to the afs_observed vector above using one of the following two approaches.
Option 1 [easy]: Plot AFS vectors for various \(N_e\) values (i.e. simulate several of them using your function
simulate_afs()), then eyeball which looks closest to the observed AFS based on the figures alone. (This is, of course, not how proper statistical inference is done, but it will be good enough for this exercie!)Option 2 [hard]: Simulate AFS vectors in steps of possible
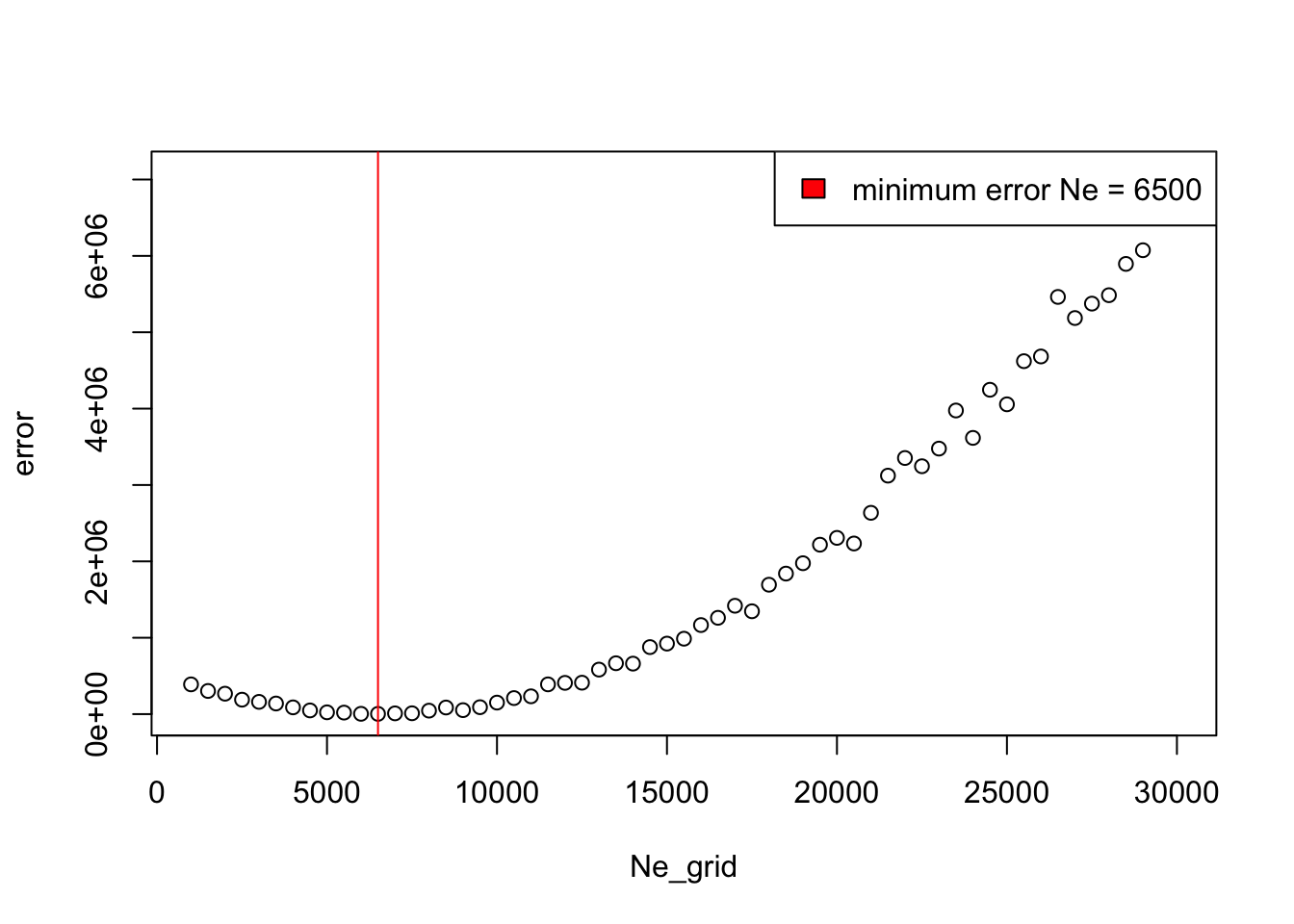
Ne(maybelapply()?), and find the \(N_e\) which gives the closest AFS to the observed AFS based on Mean squared error.
Congratulations, you now know how to infer parameters of evolutionary models using simulations! What you just did is really very similar to how simulation-based inference is done in practice (even with methods such as ABC). Hopefully you now also see how easy slendr makes it to do this (normally a rather laborious) process.
This kind of approach can be used to infer all sorts of demographic parameters, even using other summary statistics that you’ve also learned to compute… including selection parameters, which we delve into in another exercise.