import msprime
demography = msprime.Demography()
demography.add_population(name="A", initial_size=10_000)
demography.add_population(name="B", initial_size=5_000)
demography.add_population(name="C", initial_size=1_000)
demography.add_population_split(time=1000, derived=["A", "B"], ancestral="C")
ts = msprime.sim_ancestry(
sequence_length=10e6,
recombination_rate=1e-8,
samples={"A": 100, "B": 100},
demography=demography
)Why use simulations?
Many problems in population genetics cannot be solved by a mathematician, no matter how gifted. [It] is already clear that computer methods are very powerful. This is good. It […] permits people with limited mathematical knowledge to work on important problems […].
Why use simulations?
- Developing intuition into statistics
- Estimating model parameters
- Ground truth for method development
Developing intuition into statistics
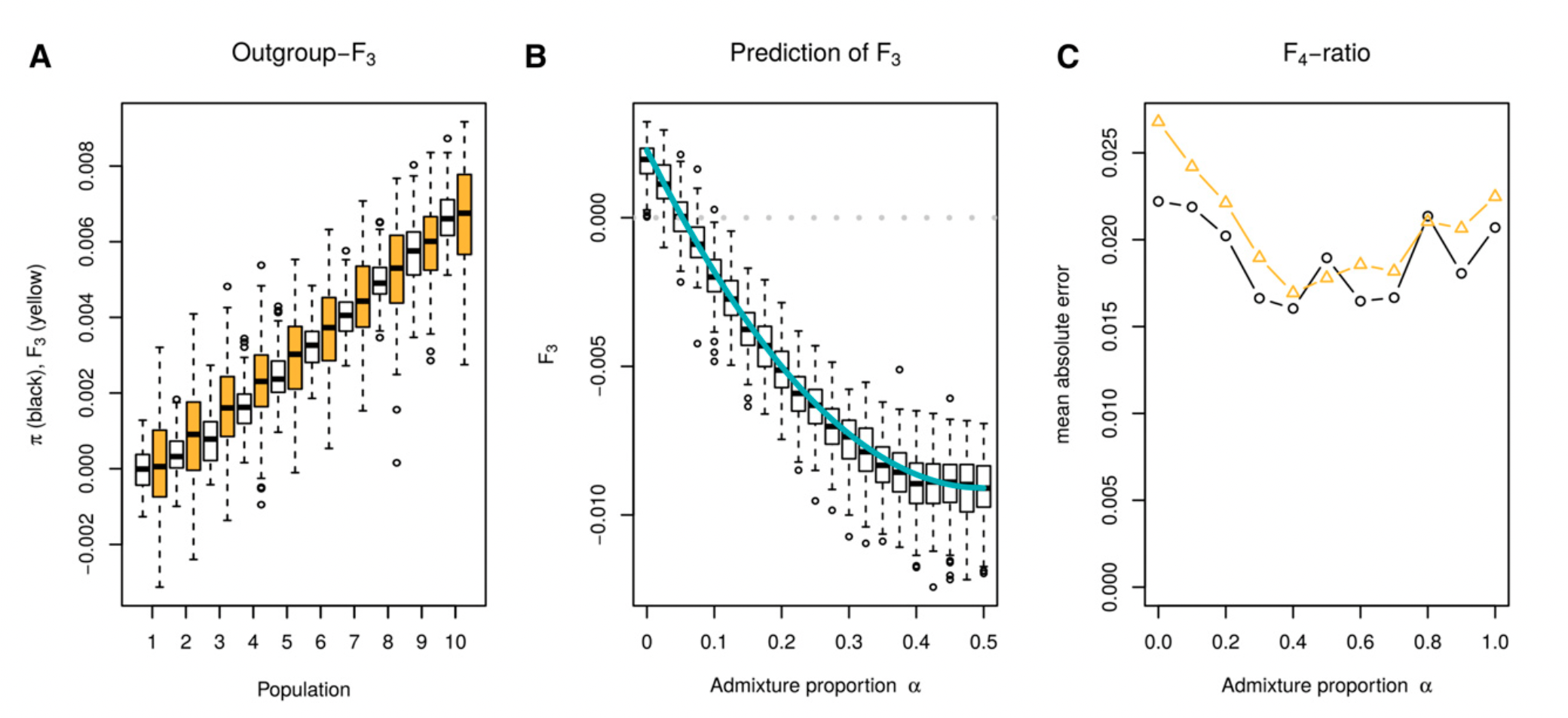
Image from Peter (2016)
Developing intuition into statistics
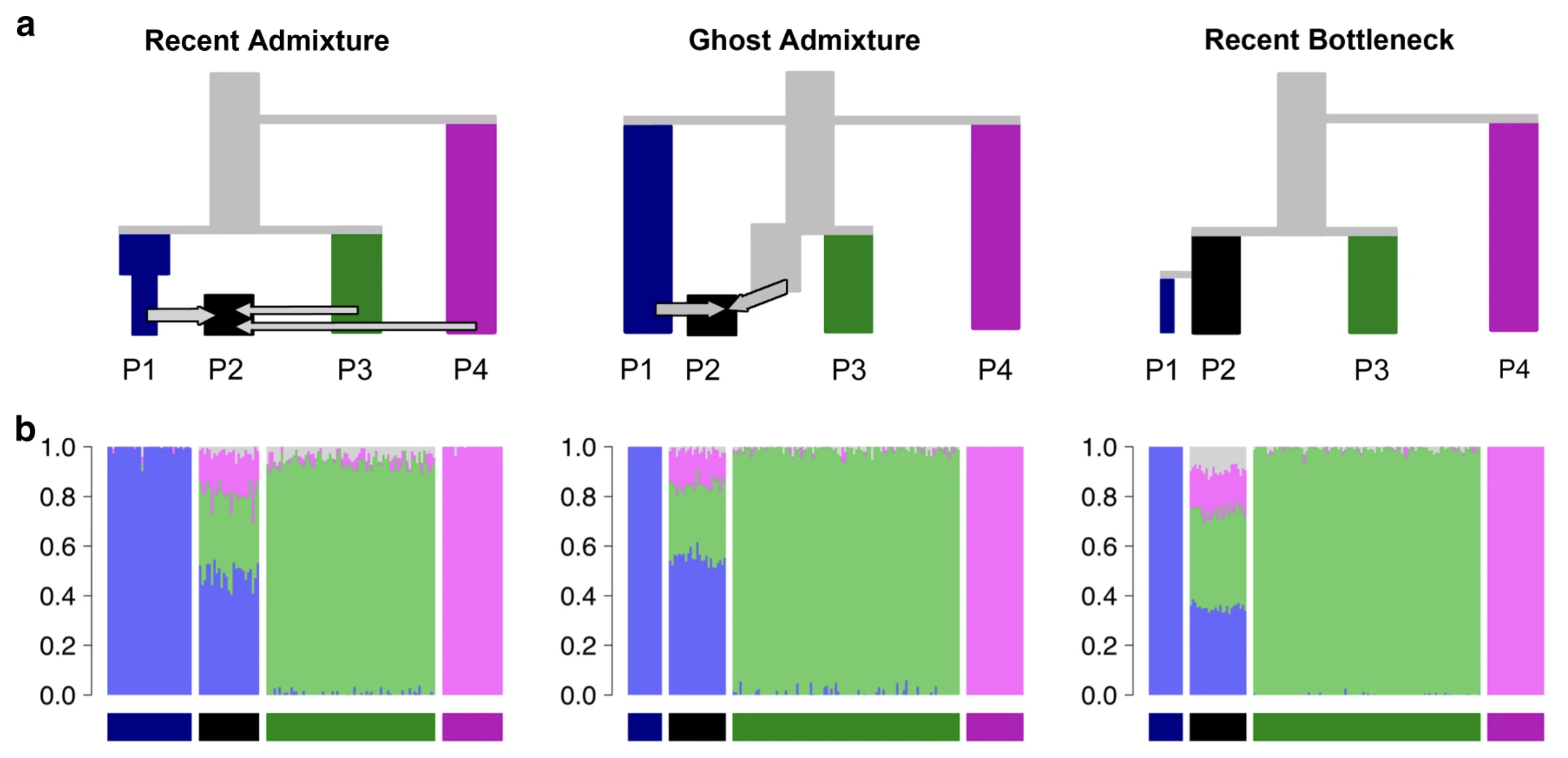
Image from Lawson et al. (2018)
Estimating model parameters (i.e. ABC)
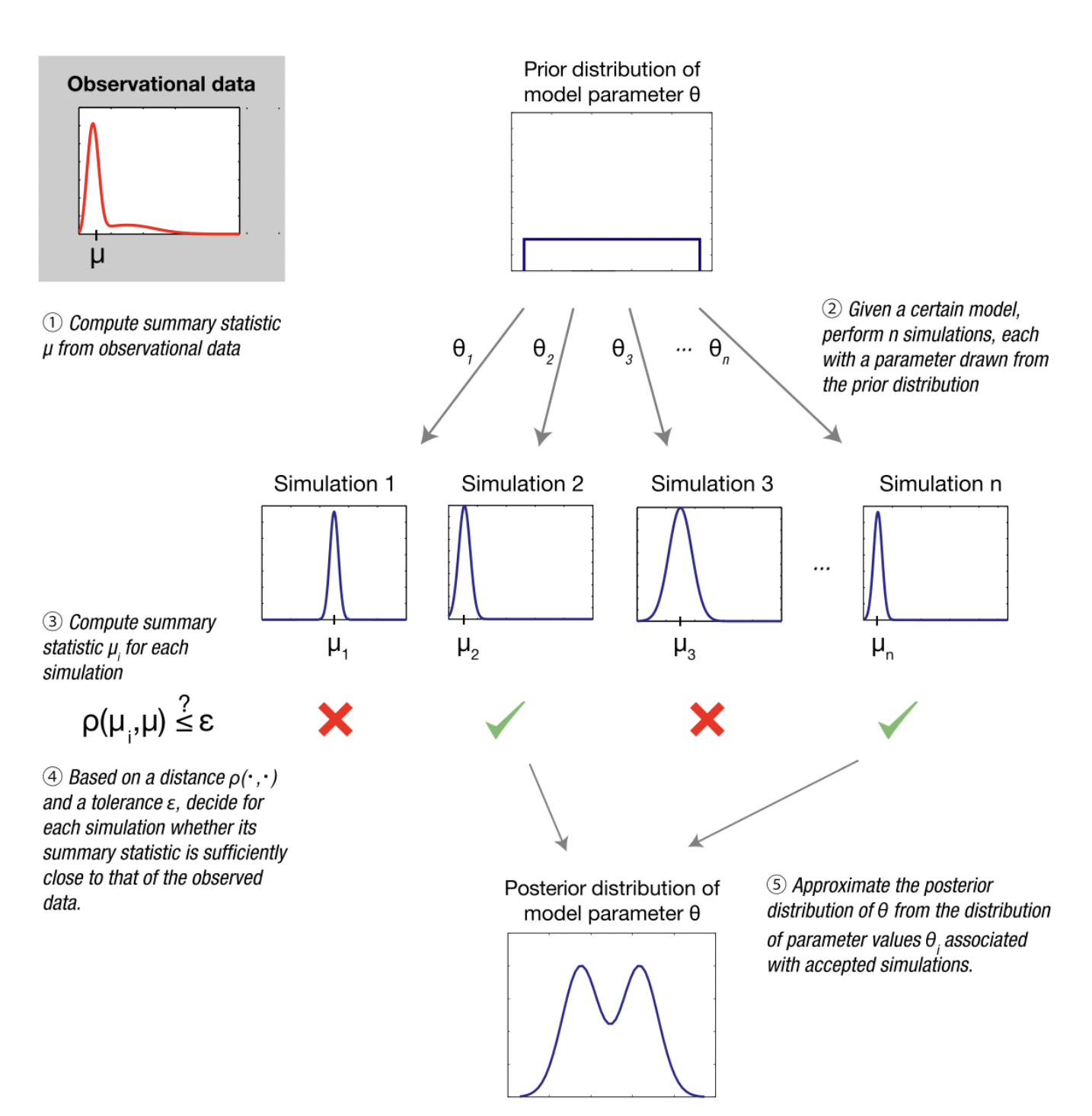
Image from Wikipedia on ABC
Ground truth for method development
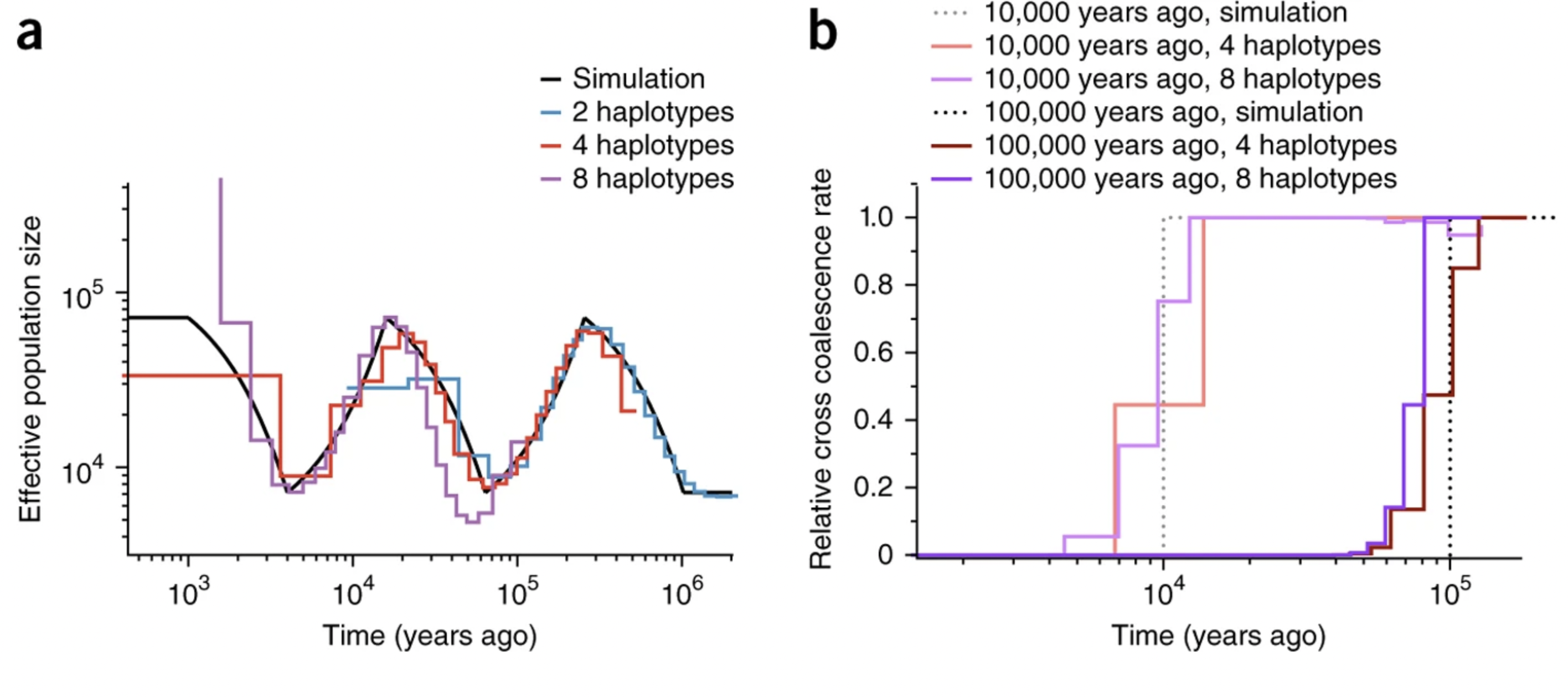
Image from Schiffels and Durbin (2014)
Simulation software
The most famous and widely used are SLiM and msprime.
They are very powerful and (nearly) infinitely flexible.
However, they both require:
- quite a bit of code for complex simulations (“complex” is relative, of course)
- relatively high confidence in programming
Our exercises will focus on the slendr
simulation toolkit for population genetics in R.
But, as a recap, let’s look at msprime and SLiM a little bit…
What is msprime?
A Python module for writing coalescent simulations
Extremely fast (genome-scale, population-scale data!)
You should know Python fairly well to build complex models
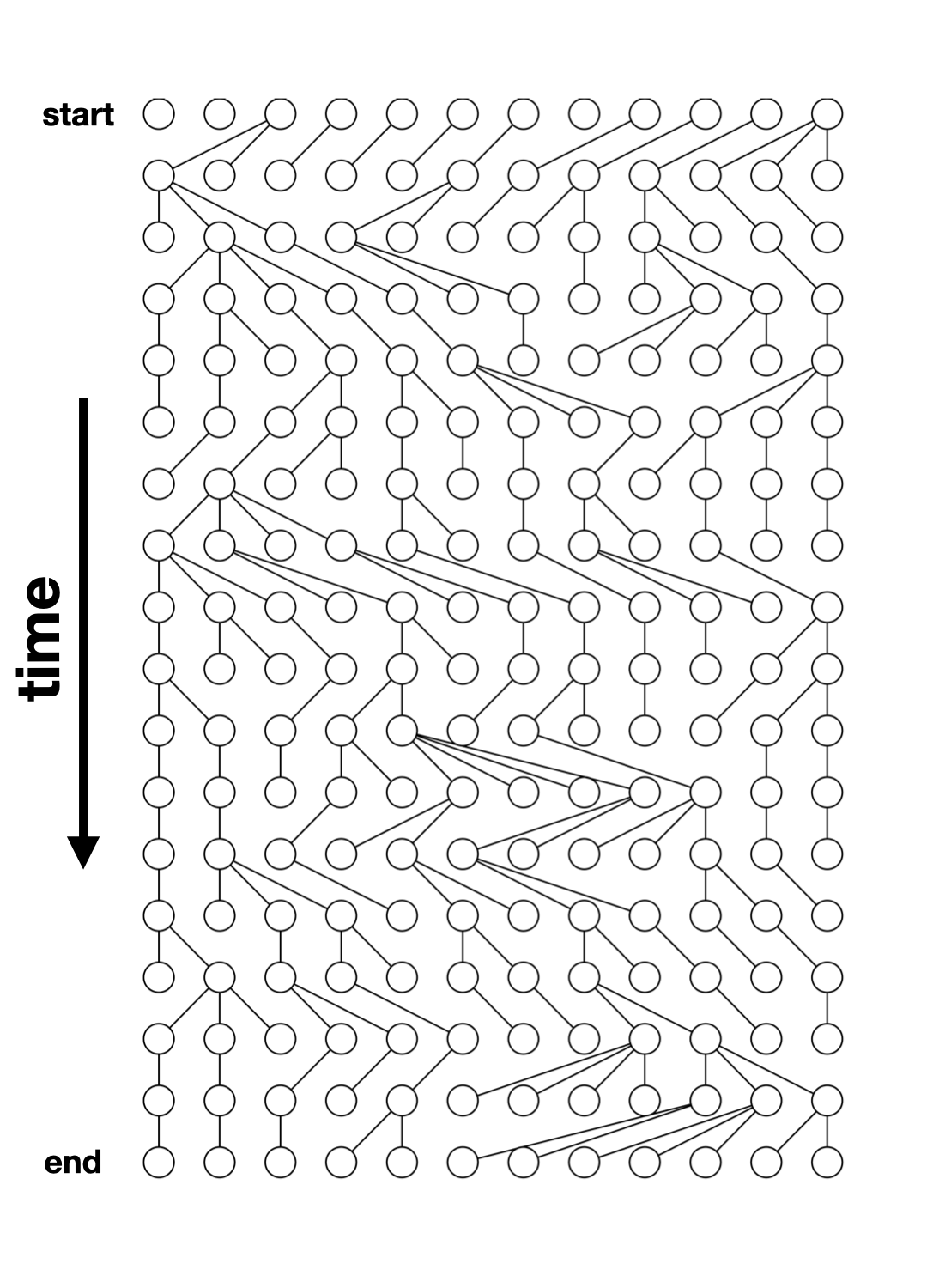
Simple simulation using msprime
What is SLiM?
A forward-time simulator
Has its own programming language
Massive library of functions for:
- demographic events
- various mating systems
- natural selection
More than 700 pages long manual!

Simple neutral simulation in SLiM
initialize() {
// create a neutral mutation type
initializeMutationType("m1", 0.5, "f", 0.0);
// initialize 1Mb segment
initializeGenomicElementType("g1", m1, 1.0);
initializeGenomicElement(g1, 0, 999999);
// set mutation rate and recombination rate of the segment
initializeMutationRate(1e-8);
initializeRecombinationRate(1e-8);
}
// create an ancestral population p1 of 10000 diploid individuals
1 early() { sim.addSubpop("p1", 10000); }
// in generation 1000, create two daughter populations p2 and p3
1000 early() {
sim.addSubpopSplit("p2", 5000, p1);
sim.addSubpopSplit("p3", 1000, p1);
}
// in generation 10000, stop the simulation and save 100 individuals
// from p2 and p3 to a VCF file
10000 late() {
p2_subset = sample(p2.individuals, 100);
p3_subset = sample(p3.individuals, 100);
c(p2_subset, p3_subset).genomes.outputVCF("/tmp/slim_output.vcf.gz");
sim.simulationFinished();
}SLiM and msprime are both incredible pieces of software…
… so why slendr?
www.slendr.net
First motivation: spatial simulations!
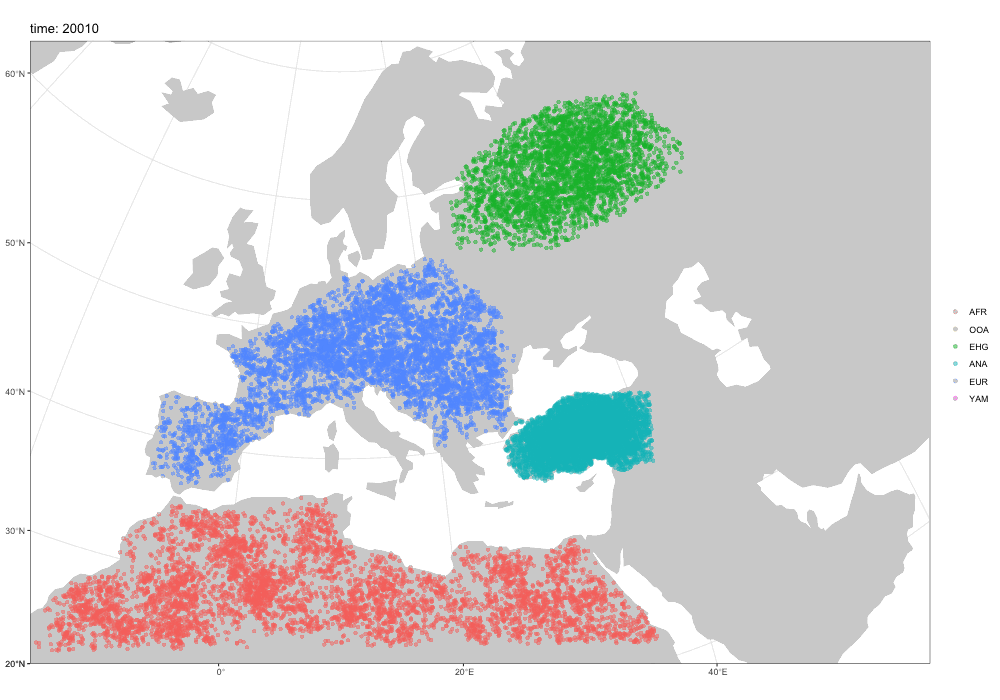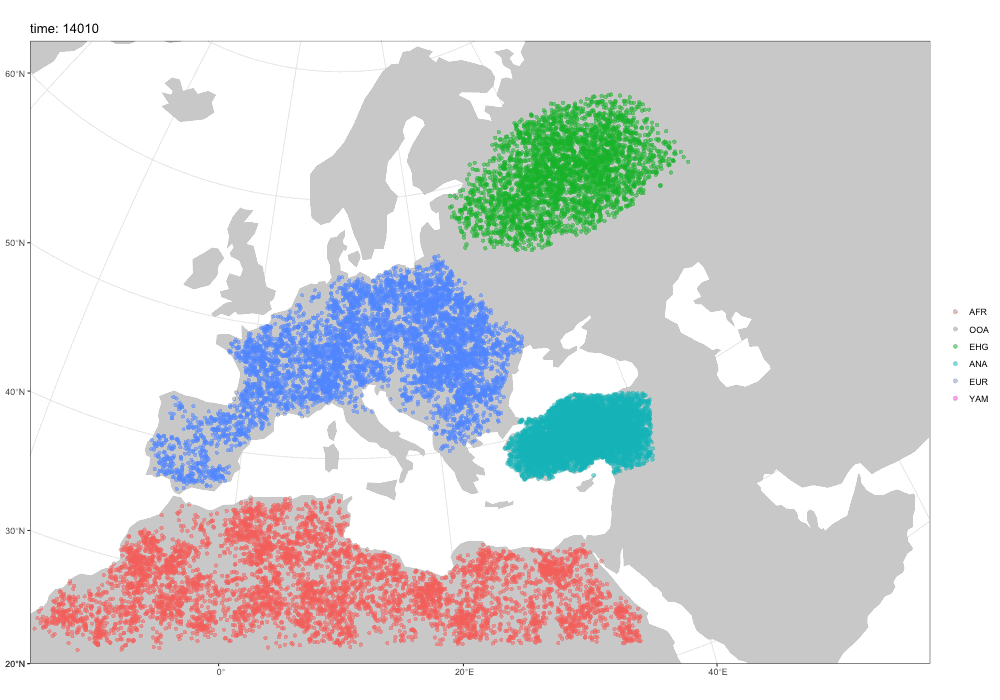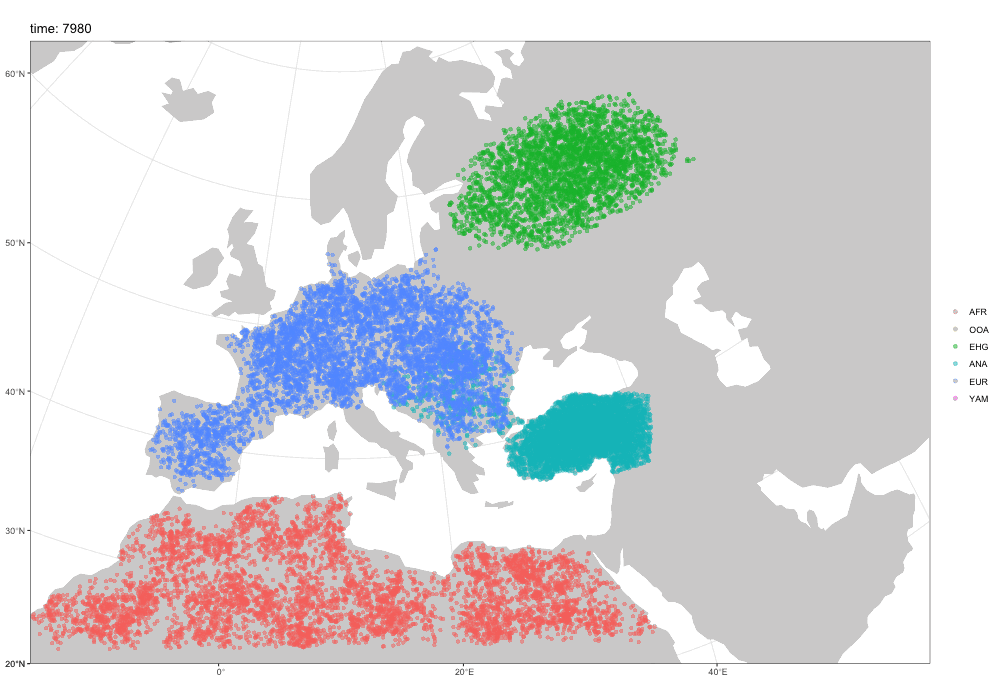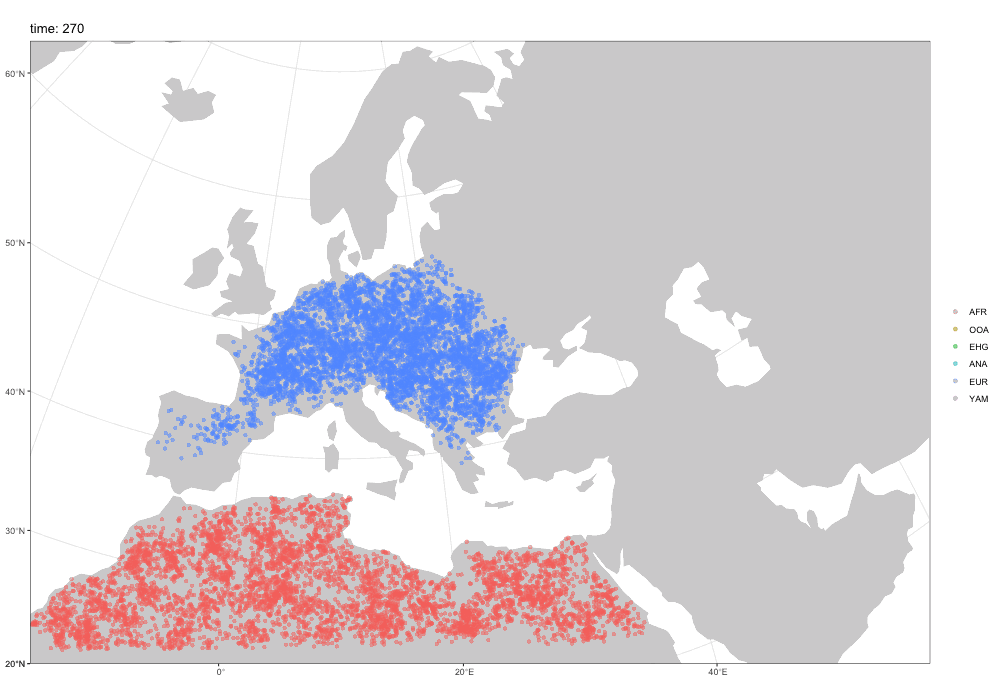
A broader motivation for slendr
Most researchers are not expert programmers
All but the most trivial simulations require lots of code
Yet, 90% [citation needed] of simulations are basically the same!
create populations (splits and \(N_e\) changes)
specify admixture rates and admixture times
… all this means duplication of code across many projects
- Computing statistics presents even more hurdles
